Building a Comprehensive Life Sciences Intelligence Platform
A startup in the pharmaceutical and biotech intelligence space embarked on a mission to create a robust platform to centralize and analyze scattered data across drug development pipelines, clinical trials, and regulatory landscapes. Sogody has partnered with them to develop and scale the platform, supporting their vision to turn complex biomedical data into actionable insights.
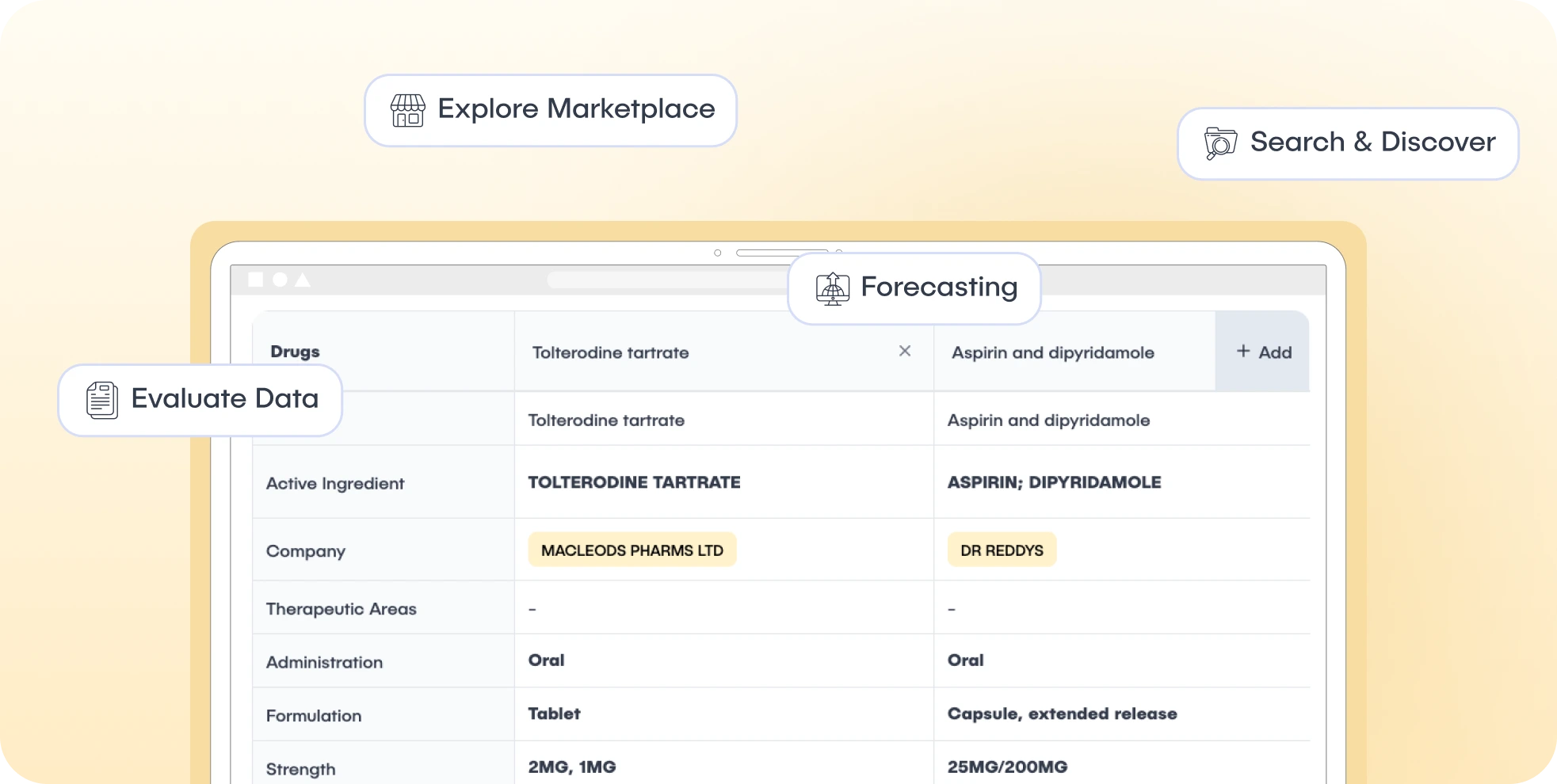
The platform is built around two core components:
1. The Data Lake
The foundation of the platform is a data lake that aggregates and harmonizes data from a broad array of sources:
Public Drug databases
Clinical trials registries
Regulatory bodies
Medical taxonomies
Our Contributions:
Platform Setup: Implemented the backend on Ruby on Rails, incorporating both manual data curation interfaces and automated workflows.
Workflow System: Engineered a custom workflow engine that orchestrates atomic steps or tasks. Each task runs in isolation on AWS Batch via Fargate containers.
Data Pipelines: Automated ingestion, transformation, and matching of data across entities like Drugs, Clinical Trials, and Regulatory submissions. The pipelines are implemented using a mix of Ruby, NodeJS, and Python, tailored to the specific requirements of each data source and processing task. Key components of the pipelines include:
Frequent Clinical Trial Syncing: Regularly ingest and update clinical trial data from registries. Post-processing includes intervention structuring, extraction, and eligibility criteria parsing. Parsed data is mapped back to internal datasets for consistency and linkage.
Regulatory Data Syncing for Drugs: Continuously pull the latest regulatory information from various regulatory sources. Information is parsed and matched back to drug entities in the platform.
Label Parsing: Extract therapeutic indications and additional metadata from regulatory drug labels, which is then mapped to diseases, drugs, and therapy requirements.
Organizational Data Enrichment: Fetch and process data on publicly traded organizations, including revenue figures and other financial and operational metrics, enriching the organizational profiles available within the platform.
AI/LLM Integration: Integrated AI and large language models (LLMs) across several stages of the data pipeline:
Named Entity Recognition (NER) and relation extraction from unstructured text sources, such as drug labels, clinical trial descriptions, and regulatory documents.
Indication Structuring: Parses therapeutic indications, links them to known diseases and drugs in the internal database, and extracts context such as required prior therapies or combination therapy use cases.
Intervention Structuring in Clinical Trials: Extracts drug names, dosages, and modes of administration from clinical trial protocols using transformer-based models.
Eligibility Criteria Structuring: Converts highly unstructured free-text eligibility criteria into structured formats for improved querying and analytics.
Data Quality Assurance: Leverages LLMs to fact-check and validate results derived in previous extraction steps, ensuring higher accuracy and reliability.
Publication Linking: Extracts publication abstracts from public sources and links them to clinical trials in the system, enriching trial records beyond standard registries.
News Processing: Scans and extracts key information from pharmaceutical news, including PDUFA dates and partnership or licensing deals between organizations.
These tasks not only parse and transform data but also link it to a structured internal schema of Drugs, Diseases, Targets, and more.
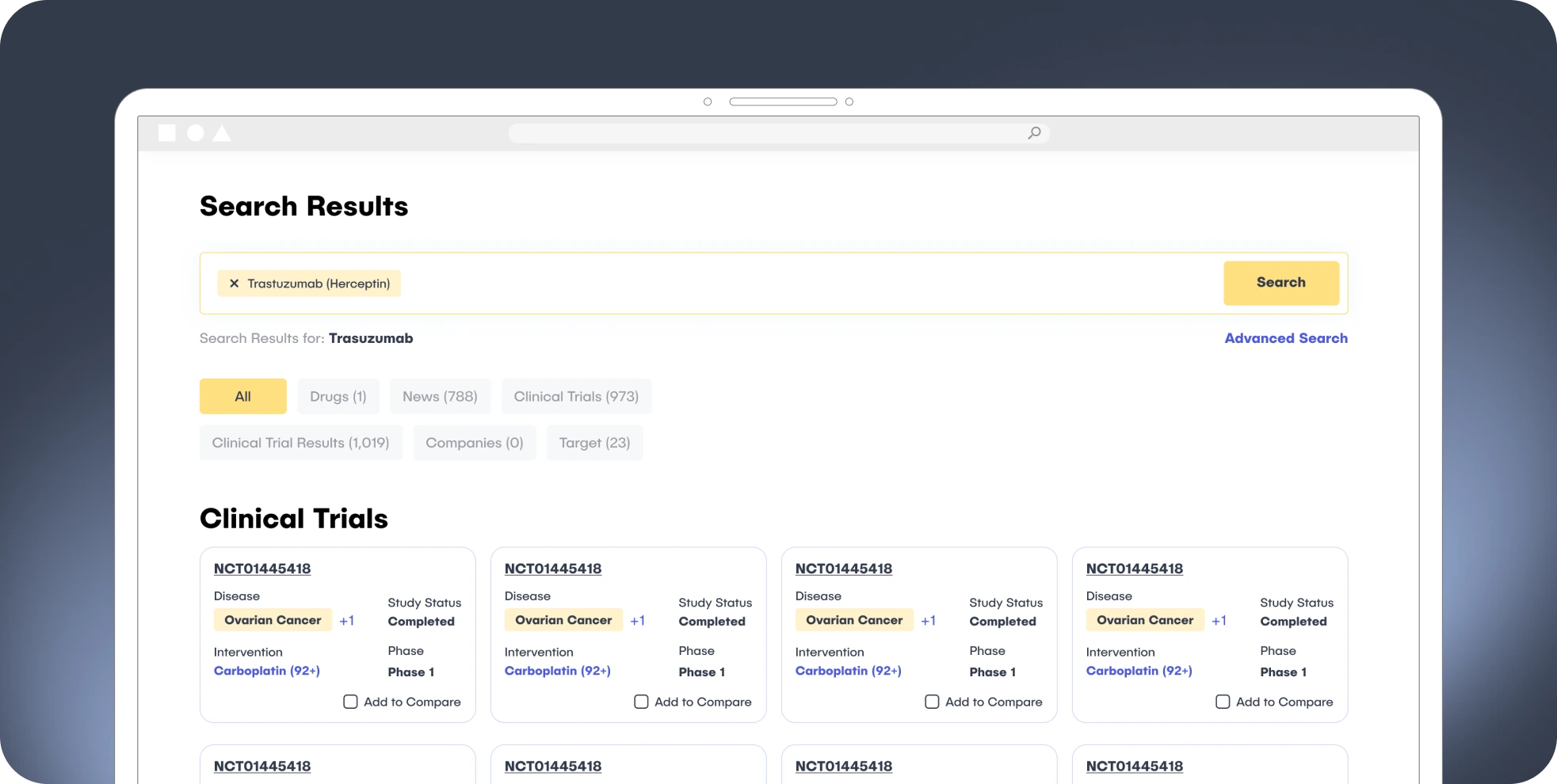
2. The Web Application
Built to serve data dynamically depending on user needs, the web application enables flexible exploration and analysis:
Key Features:
Disease-Centric View: Users can start from a disease (e.g., Breast Cancer) and access curated disease statistics, related clinical trials, and ongoing drug development.
Drug-Centric View: Allows exploration of individual drugs (by INN), detailing their development stage, associated clinical trials, and competitor analysis based on targets.
Target-Centric View: Enables users to analyze drugs, trials, and regulatory activity related to specific molecular targets.
Technology-Centric View: Provides an overview of platform technologies in drug development, showing how they are distributed across trials, diseases, and organizations.
Advanced Search: Cross-dataset search across Drugs, Trials, Targets, and Technologies with multifaceted filters.
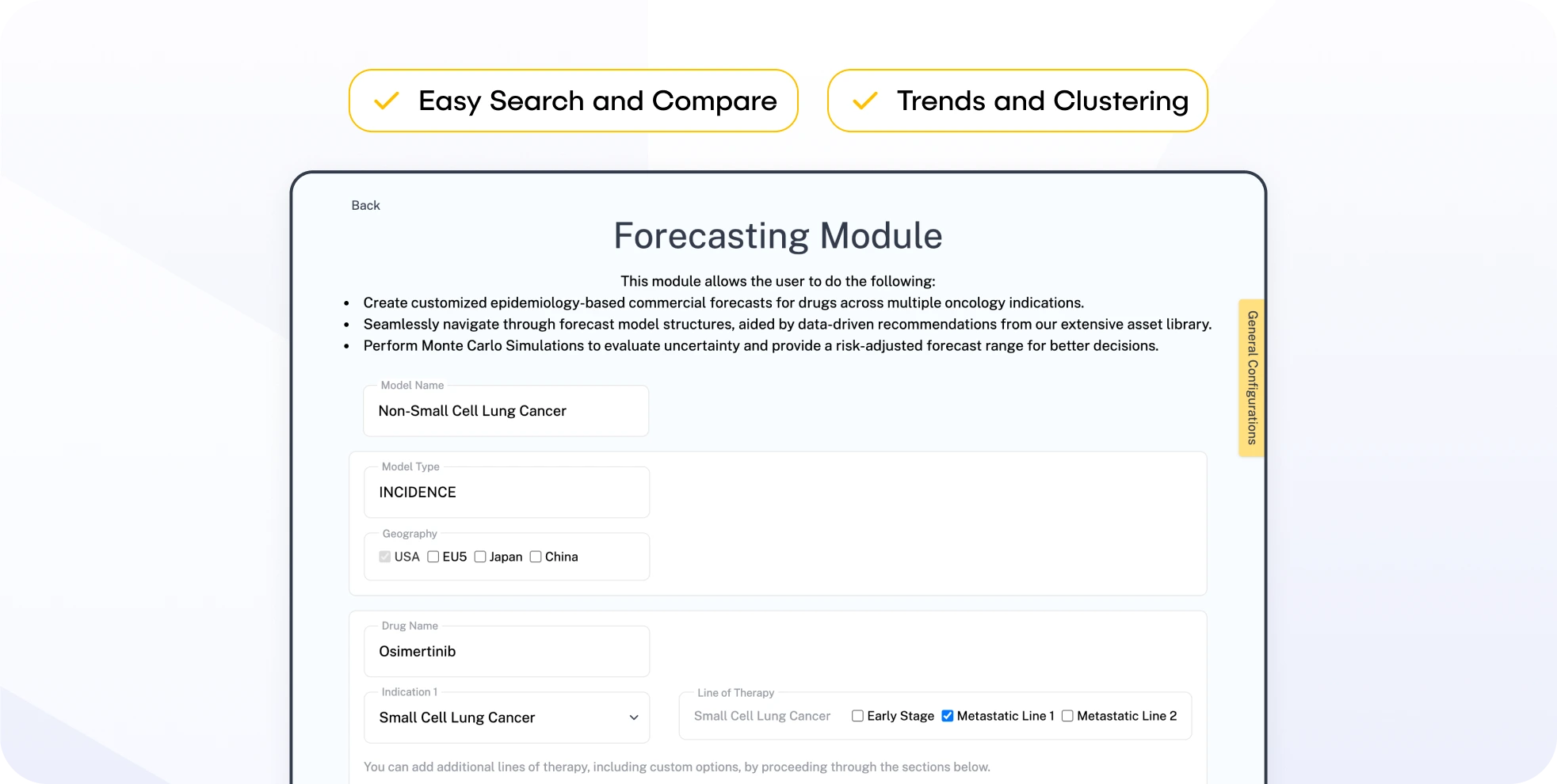
Forecasting Module: Enables users to model and compare different market scenarios using disease incidence and trial progression data. Includes persistent scenarios and Monte Carlo simulations for probabilistic forecasts.
Through this collaboration, Sogody has enabled the client to unify fragmented data sources into a coherent, AI-enriched ecosystem. The platform is now a cornerstone in strategic decision-making for pharma companies, researchers, and investment analysts.
Technologies Used
Backend: Ruby on Rails
Cloud Infrastructure: AWS (Fargate, Batch)
Data Processing: Custom workflow engine, data pipelines built with Ruby, NodeJS, and Python
AI/LLM: NLP models for text extraction, classification, indication mapping, trial structuring, fact-checking, and news/event extraction
The result is a powerful, scalable, and extensible platform that empowers users with a 360-degree view of the drug development landscape — from molecule to market.
Not there yet?
Unsure with your needs? or if you have any questions, book a call with us. we’d be happy to explore how we can create a plan that fits you perfectly.
